Lab 6
Get the code
Find the Lab 6 code in the class repository.
$ ls
README.md classes/ labs/ scripts/
$ ls labs/lab6
README cpp/ julia/ python/ rust/
$ ls labs/lab6/python
en.gp ft.gp traj.gp well.dat well.py
The instructions here focus on Python, but you may carry out the comparable steps in C++, Julia, or Rust.
Harmonic oscillator trajectory
The program well.py simulates a particle of mass \(\mathsf{m}\)
moving in a quadratic well of stiffness \(\mathsf{K}\). The time
evolution is carried out using the standard Velocity Verlet algorithm, and the
simulation runs over several multiples of the oscillator period
\(P = \mathsf{2 \pi/ \sqrt{K/m}}\). The time step is chosen
to be \(\mathsf{\Delta t} = P/200\).
The gnuplot scripts traj.gp and en.gp demonstrate the
oscillatory behaviour and confirm the conservation of energy.
$ python3 well.py > well.dat
runtime = 157.07963267948966
period = 6.283185307179586
freq = 0.15915494309189535
Running simulation...
Running simulation...
Done.
Computing discrete Fourier Transforms...
Done cos.
Done sin.
$ gnuplot -persist traj.gp
$ gnuplot -persist en.gp
Discrete Fourier transforms
The output to the file well.dat is broken into two blocks. The first
block lists the trajectory data in \(\mathsf{t}, \mathsf{x},
\mathsf{v}, \mathsf{E}\) format. The second block lists the
\(\textsf{cos}\) and \(\textsf{sin}\) discrete Fourier
transforms
(in the second and third columns) as a function of frequency (in the first
column).
$ head -n 5 well.dat
0.000000 0.999507 -0.031408 0.500000
0.031416 0.998027 -0.062785 0.500000
0.062832 0.995562 -0.094101 0.499999
0.094248 0.992114 -0.125323 0.499998
0.125664 0.987687 -0.156422 0.499997
$ tail -n 5 well.dat
31.799158 0.214164 0.001484
31.805524 0.211001 0.001170
31.811890 0.208605 0.000867
31.818256 0.206926 0.000574
31.824622 0.205932 0.000285
$ head -n 5006 well.dat | tail -n 12
156.891137 0.988677 0.150038 0.499997
156.922553 0.992903 0.118912 0.499998
156.953969 0.996149 0.087668 0.499999
156.985385 0.998411 0.056337 0.500000
157.016801 0.999689 0.024951 0.500000
157.048217 0.999979 -0.006460 0.500000
0.000000 0.205602 0.000000
0.006366 0.205932 -0.000285
0.012732 0.206926 -0.000574
0.019099 0.208605 -0.000867
The corresponding spectum is strongly peaked at the natural frequency \(\mathsf{\nu \doteq 0.159}\).
$ gnuplot -persist ft.gp
Stochastic evolution
We now want to include some random behaviour in the time evolution of the particle. In particular, we imagine that the particle is immersed in a fluid in thermal equilibrium at temperature \(T\). The fluid provides both a drag on the particle and a random stochastic force due to the fluctuating impact of the fluid molecules.
Start by building your own high-quality random number generator. Blum Blum Shub (BBS) is a famous scheme that generates one random bit per iteration. Implement BBS with the parameters \(\mathsf{p} = \mathsf{2\,888\,799\,659}`\) and \(\mathsf{q} = \mathsf{3\,694\,568\,831}\), say, so that the modulus is \(\mathsf{M} = \mathsf{pq} = \mathsf{10\,672\,869\,179\,144\,828\,629}\). The seed \(\mathsf{x_0}\) must be coprime to \(\mathsf{M}\). Let’s choose \(\mathsf{x_0} = \mathsf{326\,694\,129}\).
Write a function R() that returns a random number drawn uniformly
from the interval \(\mathsf{[0,1)}\). You’ll have to generate 52
bits (the width of the mantissa) to populate one double-precision
floating-point value.
Then write another function RN() that returns a random number drawn
from the normal (i.e., Guassian) distribution with zero mean and unit
variance. You should use the Box-Muller trick.
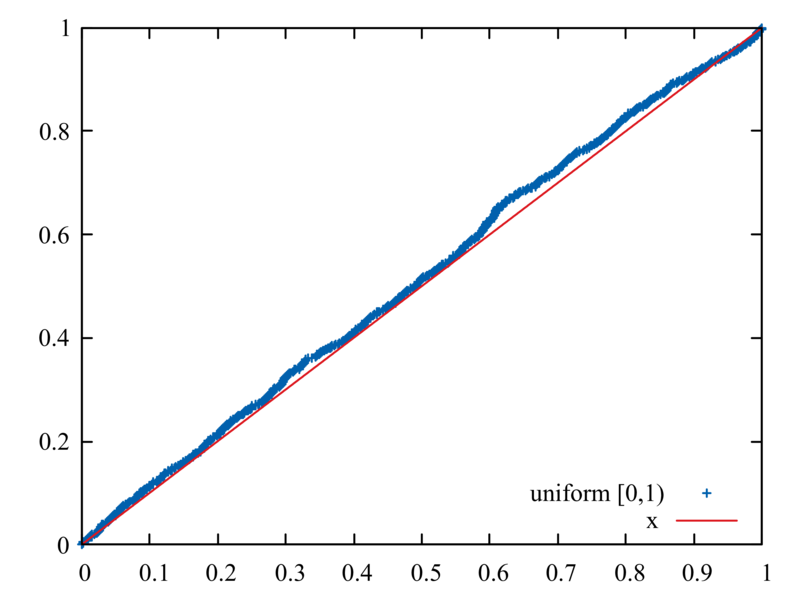
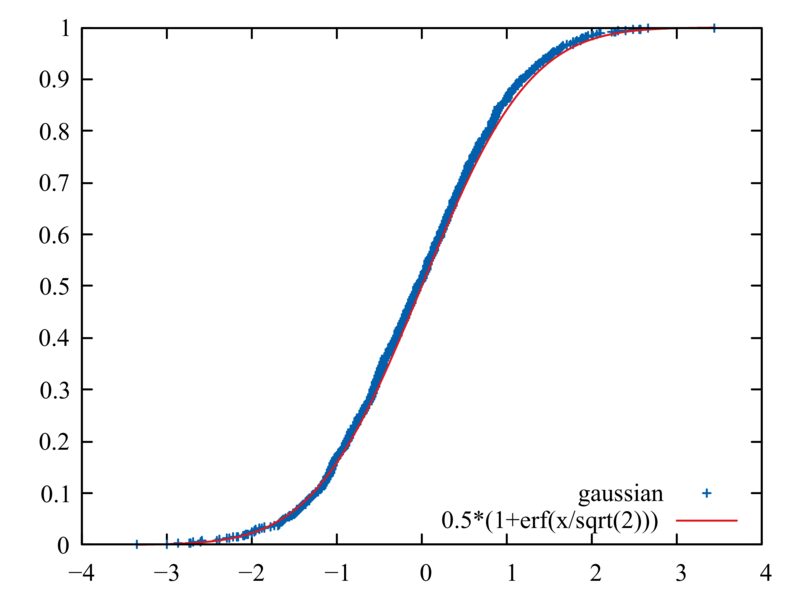
The following is a good way to test that your generators are unbiased
and performing correctly. Produce 1000 random numbers each with R()
and RN(), arranged in ascending order; compare each set to its
corresponding cumulative probability distribution
import math
import sys
import struct
print("Testing random number generator", file=sys.stderr)
r=[]
rr=[]
for n in range(1000):
r.append(R())
rr.append(RN())
r.sort()
rr.sort()
f = open("rng-test.dat","w")
for n in range(len(r)):
f.write("{:8.4} {:15.8} {:15.8}\n".format((n+0.5)/len(r),r[n],rr[n]))
f.close()
exit()
set key bottom right
plot[][0:1] "rng-test.dat" using 2:1 title "uniform [0,1)", x
pause -1
plot[][:1] "rng-test.dat" using 3:1 title "gaussian", 0.5*(1+erf(x/sqrt(2)))
pause -1
Langevin equation
Rework the simulation to accommodate an expanded set of forces, including \(\mathsf{F}_\textsf{drag} = - \mathsf{\gamma v}\) and \(\mathsf{F}_\textsf{thermal}\) in addition to the Hooke’s law force \(\mathsf{F(x)} = -\mathsf{V'(x)} = -\mathsf{Kx}\). Use the following Verlet-like update scheme:
Here, \(\mathsf{b} = \textsf{exp}(-\mathsf{\gamma \Delta t/2m})\)
and each \(\mathsf{\beta} = \sqrt{\mathsf{2 \gamma k_B T \Delta
t}}\,\xi\) is a random value with \(\xi\) representing the result of
the function call RN().
Perform several runs in which you systematically ramp up the damping from zero. The temperature can be fixed at the value \(\mathsf{k_BT} = \mathsf{1/2}\), consistent with initial conditions and the energy predicted by equipartition. Make a plot showing the resulting changes in the Fourier transform peak.